| oligos_5-8nt_m1_shift9 (oligos_5-8nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m1; m=0 (reference); ncol1=13; shift=9; ncol=22; ---------waAAAGTCAAAww
; Alignment reference
a 0 0 0 0 0 0 0 0 0 133 160 337 409 428 39 22 20 390 408 375 148 135
c 0 0 0 0 0 0 0 0 0 80 103 45 8 9 24 15 392 21 5 18 68 89
g 0 0 0 0 0 0 0 0 0 79 92 32 10 1 353 13 21 9 10 25 103 91
t 0 0 0 0 0 0 0 0 0 152 89 30 17 6 28 394 11 24 21 26 125 129
|
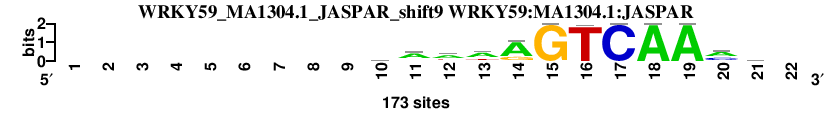
| WRKY59_MA1304.1_JASPAR_shift9 (WRKY59:MA1304.1:JASPAR) |
 |
0.916 |
0.846 |
6.359 |
0.947 |
0.967 |
3 |
1 |
29 |
3 |
3 |
7.800 |
1 |
; oligos_5-8nt_m1 versus WRKY59_MA1304.1_JASPAR (WRKY59:MA1304.1:JASPAR); m=1/166; ncol2=12; w=12; offset=0; strand=D; shift=9; score= 7.8; ---------mawwAGTCAAmr-
; cor=0.916; Ncor=0.846; logoDP=6.359; NsEucl=0.947; NSW=0.967; rcor=3; rNcor=1; rlogoDP=29; rNsEucl=3; rNSW=3; rank_mean=7.800; match_rank=1
a 0 0 0 0 0 0 0 0 0 56 109 97 103 130 0 0 0 173 173 93 46 0
c 0 0 0 0 0 0 0 0 0 48 12 15 18 0 0 2 173 0 0 62 38 0
g 0 0 0 0 0 0 0 0 0 26 25 16 8 39 173 0 0 0 0 6 58 0
t 0 0 0 0 0 0 0 0 0 43 27 45 44 4 0 171 0 0 0 12 31 0
|
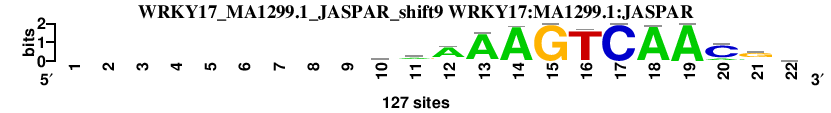
| WRKY17_MA1299.1_JASPAR_shift9 (WRKY17:MA1299.1:JASPAR) |
 |
0.873 |
0.810 |
7.729 |
0.935 |
0.945 |
22 |
4 |
6 |
10 |
18 |
12.000 |
6 |
; oligos_5-8nt_m1 versus WRKY17_MA1299.1_JASPAR (WRKY17:MA1299.1:JASPAR); m=6/166; ncol2=14; w=13; offset=0; strand=D; shift=9; score= 12; ---------aaAAAGTCAACgv
; cor=0.873; Ncor=0.810; logoDP=7.729; NsEucl=0.935; NSW=0.945; rcor=22; rNcor=4; rlogoDP=6; rNsEucl=10; rNSW=18; rank_mean=12.000; match_rank=6
a 0 0 0 0 0 0 0 0 0 57 67 94 117 125 0 0 0 125 127 20 29 32
c 0 0 0 0 0 0 0 0 0 25 24 9 0 0 0 5 127 0 0 97 9 38
g 0 0 0 0 0 0 0 0 0 23 14 18 6 1 127 1 0 2 0 8 78 32
t 0 0 0 0 0 0 0 0 0 22 22 6 4 1 0 121 0 0 0 2 11 25
|
| WRKY17.ampDAP_M0837_AthalianaCistrome_shift9 (WRKY17.ampDAP:M0837:AthalianaCistrome) |
 |
0.833 |
0.833 |
8.362 |
0.921 |
0.918 |
42 |
3 |
1 |
37 |
57 |
28.000 |
18 |
; oligos_5-8nt_m1 versus WRKY17.ampDAP_M0837_AthalianaCistrome (WRKY17.ampDAP:M0837:AthalianaCistrome); m=18/166; ncol2=13; w=13; offset=0; strand=D; shift=9; score= 28; ---------aaAAAGTCAACGm
; cor=0.833; Ncor=0.833; logoDP=8.362; NsEucl=0.921; NSW=0.918; rcor=42; rNcor=3; rlogoDP=1; rNsEucl=37; rNSW=57; rank_mean=28.000; match_rank=18
a 0 0 0 0 0 0 0 0 0 172 246 353 383 386 0 0 0 389 388 29 3 110
c 0 0 0 0 0 0 0 0 0 69 66 3 0 0 0 20 389 0 0 359 35 118
g 0 0 0 0 0 0 0 0 0 62 44 22 1 3 389 0 0 0 0 0 305 80
t 0 0 0 0 0 0 0 0 0 86 33 11 5 0 0 369 0 0 1 1 46 81
|
| WRKY45.DAP_M0805_AthalianaCistrome_rc_shift9 (WRKY45.DAP:M0805:AthalianaCistrome_rc) |
 |
0.911 |
0.841 |
0.497 |
0.939 |
0.956 |
5 |
2 |
149 |
7 |
8 |
34.200 |
24 |
; oligos_5-8nt_m1 versus WRKY45.DAP_M0805_AthalianaCistrome_rc (WRKY45.DAP:M0805:AthalianaCistrome_rc); m=24/166; ncol2=12; w=12; offset=0; strand=R; shift=9; score= 34.2; ---------aaAAAGTCAAmg-
; cor=0.911; Ncor=0.841; logoDP=0.497; NsEucl=0.939; NSW=0.956; rcor=5; rNcor=2; rlogoDP=149; rNsEucl=7; rNSW=8; rank_mean=34.200; match_rank=24
a 0 0 0 0 0 0 0 0 0 272 391 533 558 583 0 0 0 600 597 182 138 0
c 0 0 0 0 0 0 0 0 0 130 82 15 8 0 0 1 599 0 0 383 74 0
g 0 0 0 0 0 0 0 0 0 92 69 25 8 16 600 0 1 0 2 1 297 0
t 0 0 0 0 0 0 0 0 0 106 58 27 26 1 0 599 0 0 1 34 91 0
|




